Pangenome analysis
Overview
Teaching: 10 min
Exercises: 50 minQuestions
How to determine a pangenome from a collection of isolate genome sequences?
Objectives
Interpret the output of a pangenome analysis.
Pangenome analysis
The microbial pangenome is the union of genes shared by genomes of interest. The core genome is the intersection of genes of these genomes, thus core genes are genes present in all strains. The accessory genome (also: variable, flexible, dispensable genome) refers to genes not present in all strains of a species. These include genes present in two or more strains or even genes unique to a single strain. Acquired antibiotic resistance genes are typically genes of the accessory genome.
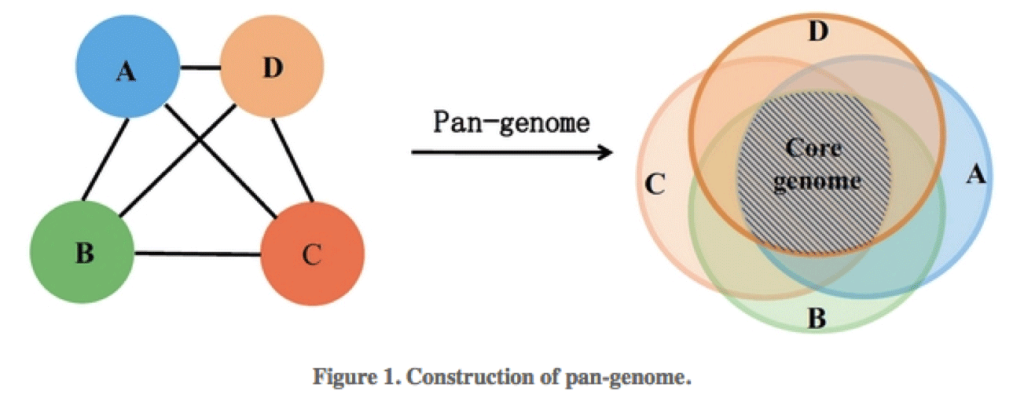
Find more information of the pangenome here
Extract genes recX, wzx and dnaA from two genomes.
For this, we need to find if the genes are present in the annotated files.
Challenge: How can I extract genes from the file with the genes?
Hints:
grep -A 10 'genename' *.gbk #gives you the first ten line after the gene name was found. Remember that grep is case sensitive!Solution
$ cd ~/annnotation $ grep -A 10 'recX' */*gbk $ grep -A 10 'RecX' */*gbk
Note for each genome if the gene has been found or not. Mark them as present (1) in the Google Sheet, else mark the gene that differs or is absent as 0.
Pangenome analysis
Comparing every gene of every genome to every other gene in the dataset is an enourmous task, and takes a long time even if automated. Roary is a pipeline to determine genes of the core and pangenome. It takes a few short-cuts such as clustering instead of pair-wise alignment and can perform this task in a relatively short time frame. An excellent step-by-step tutorial can be found here
First we need to find the files generated by prokka. We are using the annotations prepared beforehand for all genomes and we will copy them from the shared folder. How many are there?
$ cd ~
$ cp /mnt/netappits/users/courses2/shared/triumph/gff.zip ~/gff.zip
$ unzip gff.zip
$ ls ~/gff/*.gff
Then we can go to your home folder again and start roary. The -s option tells it to disregard genetic context, as we are looking at draft assemblies with different contigs and they are not ordered, the -r option generates some interesting plots in R. The 1> and 2> redirect the standard screen output and the error output to a file for later viewing. The -f option tells it where to store the output files (the folder orthology). The -i 80 options tells Roary to use a sequence identity of 80% when clustering the proteins. The -e -n option tells it to generate a core gene alignment of all core genes (this is slow. consider not using this as it takes overnight to run). The -p 2 option tells it to use 2 CPUs. Roary needs quite some CPU power. We are using the 1> and 2> redirects to store the screen outputs as it generates a lot of noise. 1> redirects the standard output, 2> redirects the error output.
$ conda activate roary
$ cd ~
$ roary ~/gff/*.gff -s -r -f orthology_en -i 80 -p 2 -e -n >roary.stdout.log 2> roary.error.log # This might be slow.
Discussion: Open or closed pangenome?
After roary finishes, have a look at the summary file in the output folder. How many core and pangenome genes are there? Visit the definition of an open and closed pangenome here. Does E. coli have a closed or an open pangenome? View the file Rplots.pdf or the file conserved_vs_total_genes.png in your browser to have a different visualization of the core and pan genome.
Visualization
Some genes are present in all genomes, some are present in some and absent in others. Data on presence and absence of genes was collected in a matrix called gene_presence_absence.csv. Clustering of this information was used to build a tree (available as accessory_binary_genes.fa.newick). As a next step we are going to visualize this clustering.
Challenge: Which isolate(s) is/are related to genome barcode02 based on gene presence/absence??
Copy accessory_binary_genes.fa.newick and gene_presence_absence.csv to your own computer. Open phandango in Chrome, drop file accessory_binary_genes.fa.newick and then file gene_absence_presence.csv.
Download the accessory_binary_genes.fa.newick and gene_absence_presence.csv files using your webbrowser. Drag and drop them into the phandango website. Look at the isolates that are closest to barcode02 in your tree.Solution
x strains are closely related .. .. ..
Key Points
The microbial pangenome is the union of genes in genomes of interest.
The microbial core genome is the intersection of genes shared by genomes of interest.
Roary is a pipeline to determine genes of the pangenome.